Software for drug design and research
Powerful interactive tools for exploring biological systems.
Molchanica Suite for structural biology
A single toolchain for viewing, building, simulating, and validating macromolecules. Molchanica combines capabilities found in PyMOL, Coot, VMD, and MD packages, with GPU-accelerated Rust backends and modern visualization.
Molchanica
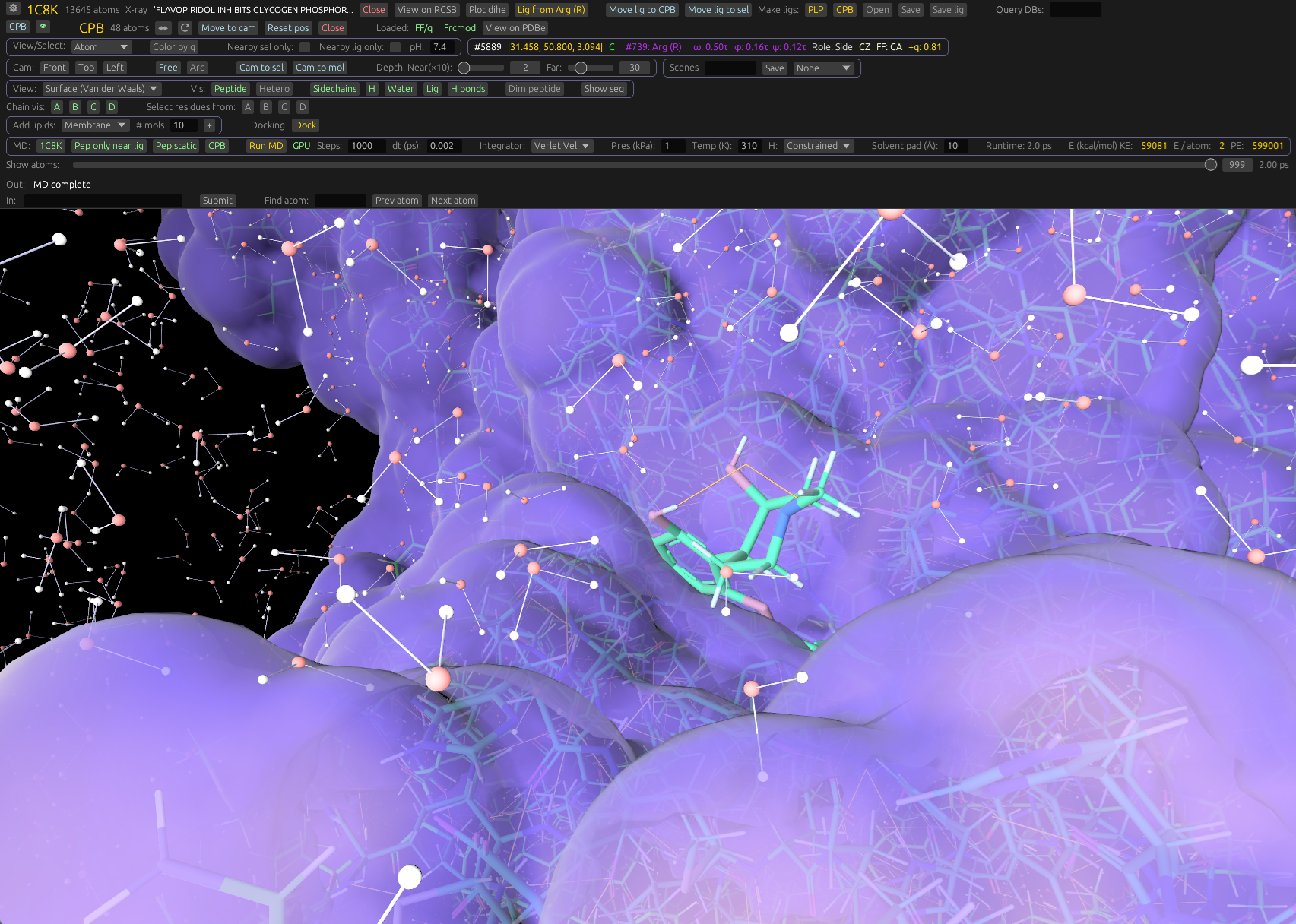
View and edit molecules, easy-to-use molecular dynamics, and docking.View and manipulate proteins, organic molecules, lipids, nucleic acids, and electron density. Integrated dynamics. Fast on modern GPUs.
Details and downloadPlasCAD
Plasmid design and validationDesign plasmids, validate primers, and keep small molecular biology workflows organized.
Details and downloadRust and Python structural bio libraries
Foundational bio libraries in RustOperate on common file formats, and interact with database APIs. Run MD with Amber parameters. Includes marching cubes isosurfaces and more.
Details and downloadMolchanica
View proteins and other molecules in 3D, render electron density, run molecular dynamics, and perform docking. Built on a modern Rust engine.
DownloadPlasCAD
Visual plasmid editor for day-to-day molecular biology. Primer design, feature annotation, and validation tools.
DownloadBio libraries for Rust and Python
Foundational crates and bindings that make structural biology native to modern languages.
Why Athanor
We make powerful software for scientists. Everything is open-source, fast, and easy to use.
The software just works. A smooth user experience is our priority.