PlasCAD
Design plasmids, generate primers, validate quickly
A fast desktop tool for primer tuning and cloning workflows (SLIC/FastCloning), with WYSIWYG sequence editing, circular maps, restriction analysis, and interoperability with GenBank/SnapGene/FASTA.
Why PlasCAD
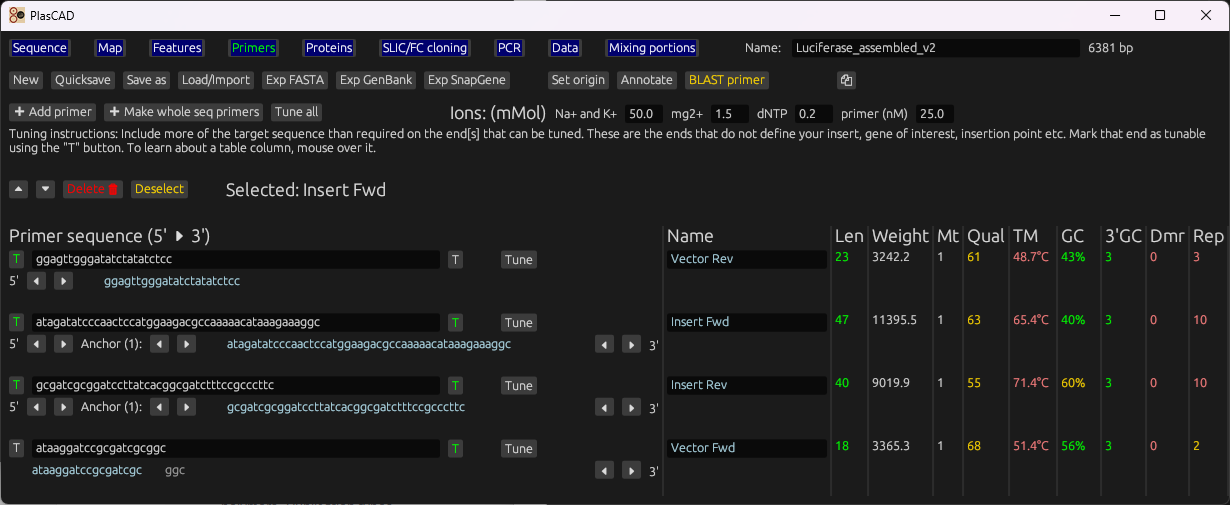
Primer QC and tuning
Nearest-neighbor Tm with salt compensation, GC%, 3′ stability, repeats, and dimer checks. Auto-extend ends to optimize.
Cloning-ready
Generate SLIC/FastCloning primers from vector/insert + site, create product files, and validate designs.
See everything
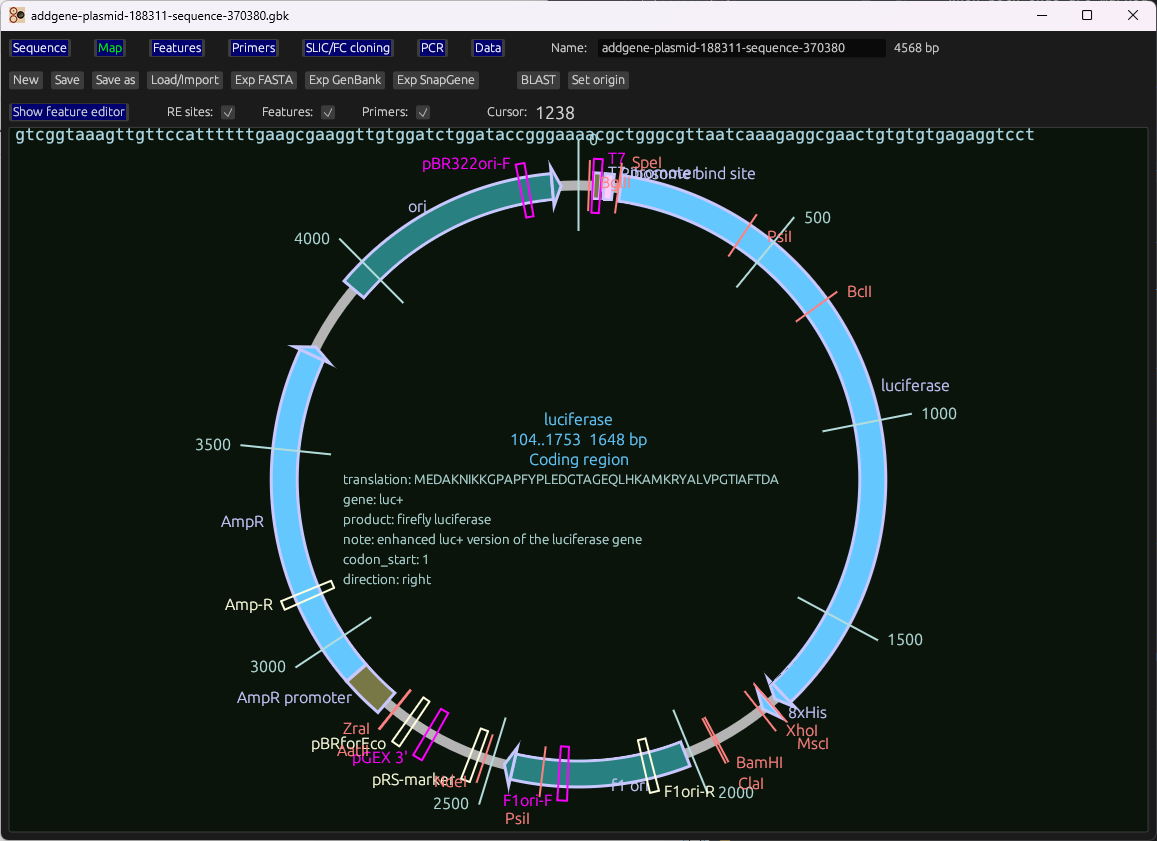
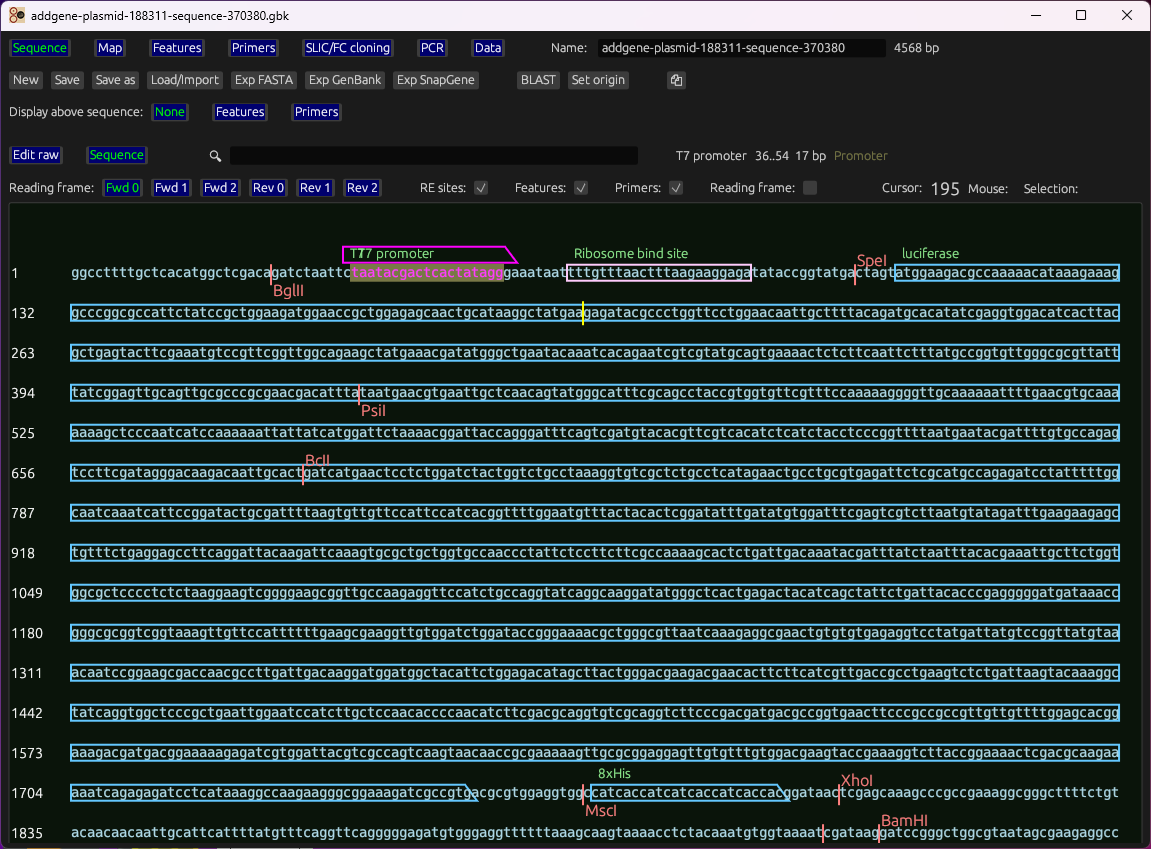
Linear editor and circular map with features, primers, restriction sites, ORFs, protein view, hydropathy plots.
Plays nice with others
Read/Write GenBank; read SnapGene .dna and FASTA; AB1 trace viewer; automatic feature annotation for common motifs.
What you can do in minutes
1. Open or import
Drag in GenBank/FASTA/SnapGene or start fresh. Common features auto-annotate on import.
2. Design primers
Tune ends until Tm and quality metrics meet targets; generate SLIC/FC primers with overlaps.
3. Validate and export
Check restriction sites, ORFs, hydropathy; export GenBank or native .pcad for compact storage.
Gallery
Representative views from the README
For technical users
File formats
Read: GenBank, FASTA, SnapGene .dna, AB1. Write: GenBank, FASTA; native .pcad (compact, includes features/primers).
Primer calculations
SantaLucia/Hicks NN model with ion corrections; quality as weighted metric for auto-tuning.
Annotations
Dynamic primer binding, common restriction enzymes, tags (e.g., His6/8), and automatic feature detection.
Protein context
AA translation per ORF, hydropathy plots, optional PDB lookup for matching coding regions.
See the repository README for full details, AB1 notes, near-term roadmap, and algorithm references.