Integration with online databases
This application integrates with the following online databases. This allows searching by molecule name or ID, downloading molecules, and discovering related ones.
Load molecules from identifier or name
The most basic demonstration of this is to type a molecule's identifier or common name into the Query DBs field near the top of the window, and either press the Enter key, or click the query button associated with the desired database. The buttons selectively appear based on the text typed. For example, if the query starts with "DB", the "Load DrugBank" button will appear. If it's at least 4 characters long, "Load from RCSB" appears. (This supports both historic 4-character identifiers, and the newer 12-character system. ).
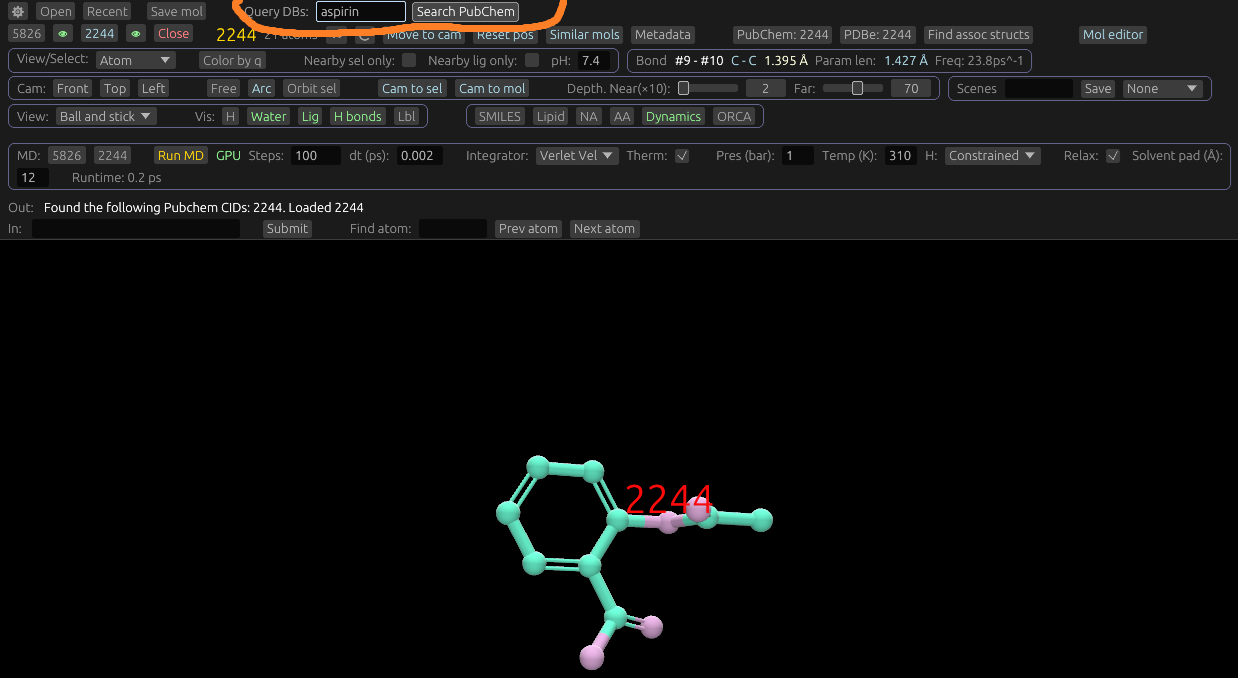
If you type the name of a molecule. (For example: "aspirin"), "Search PubChem" will appear. It will open the best match, if at least one molecule is found. Other matches will be displayed in the Out portion.
When you click one of these buttons, the application will attempt to load the molecule with that identifier or name. If unable to load (Network or database problem, invalid identifier etc), an error message will be displayed under Out.
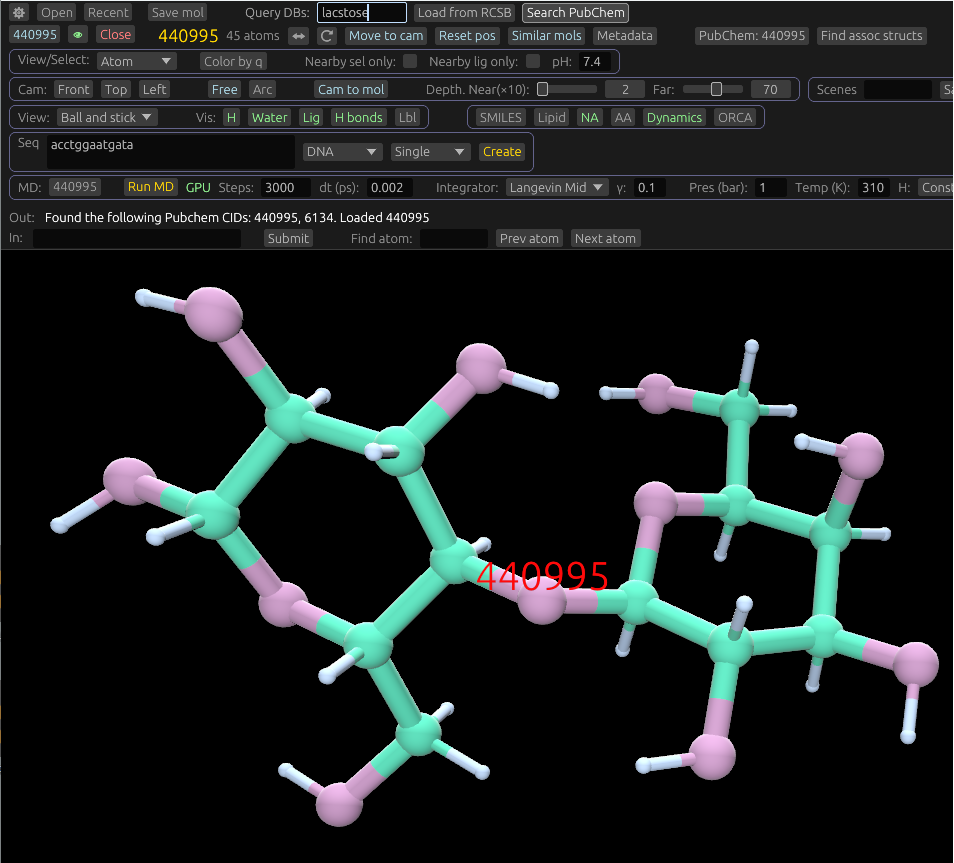
For example, try searching for "aspirin", "lactose", "glucose", "cylclbutane", "benzocaine", or other named molecules using the pubchem search. Sugars, hormones, carbohydrates, and drugs are all things that can be searched for.
Open the database page in a web browser
Once a molecule is opened, buttons will appear on the molecule's GUI line with database names. Clicking one will open your operating system's default web browser to that database's page for the molecule. You will find detailed information on it there.
Find associated structures
Click the "Find assoc structs" button in the molecule's section to query PubChem for associated structures. For proteins, this is often ligands that are known to bind to the protein.